Modeling intracellular spatial heterogeneity in bacterial cells
Intracellular spatial heterogeneity is frequently observed in bacteria, where the chromosome occupies part of the cell’s volume and a circuit’s DNA often localizes within the cell. How this heterogeneity affects core processes and its implications to genetic circuit is still poorly understood. In fact, commonly used ordinary differential equation (ODE) models of genetic circuits assume a well-mixed ensemble of molecules and, as such, do not capture spatial aspects. Reaction-diffusion partial differential equation (PDE) models have been only occasionally used since they are difficult to integrate and do not provide mechanistic understanding of the effects of spatial heterogeneity. My research uses tools from dynamical systems theory such as timescale sepertion and contraction theory to derive reduced ODE model's that capture spatial effects, yet have the same dimensionality as commonly used well-mixed models. In particular, the only difference with respect to a well-mixed ODE model is that the association rate constant of binding reactions is multiplied by a coefficient, which we refer to as the binding correction factor (BCF). The BCF depends on spatial parameters such as the size of interacting molecules and on their location when fixed in space and it is equal to unity in a well-mixed ODE model. The BCF can be used to investigate how spatial heterogeneity affects the behavior of core processes and genetic circuits. This simple-to-use model can be used to both analyze and design genetic circuits while accounting for spatial intracellular effects.
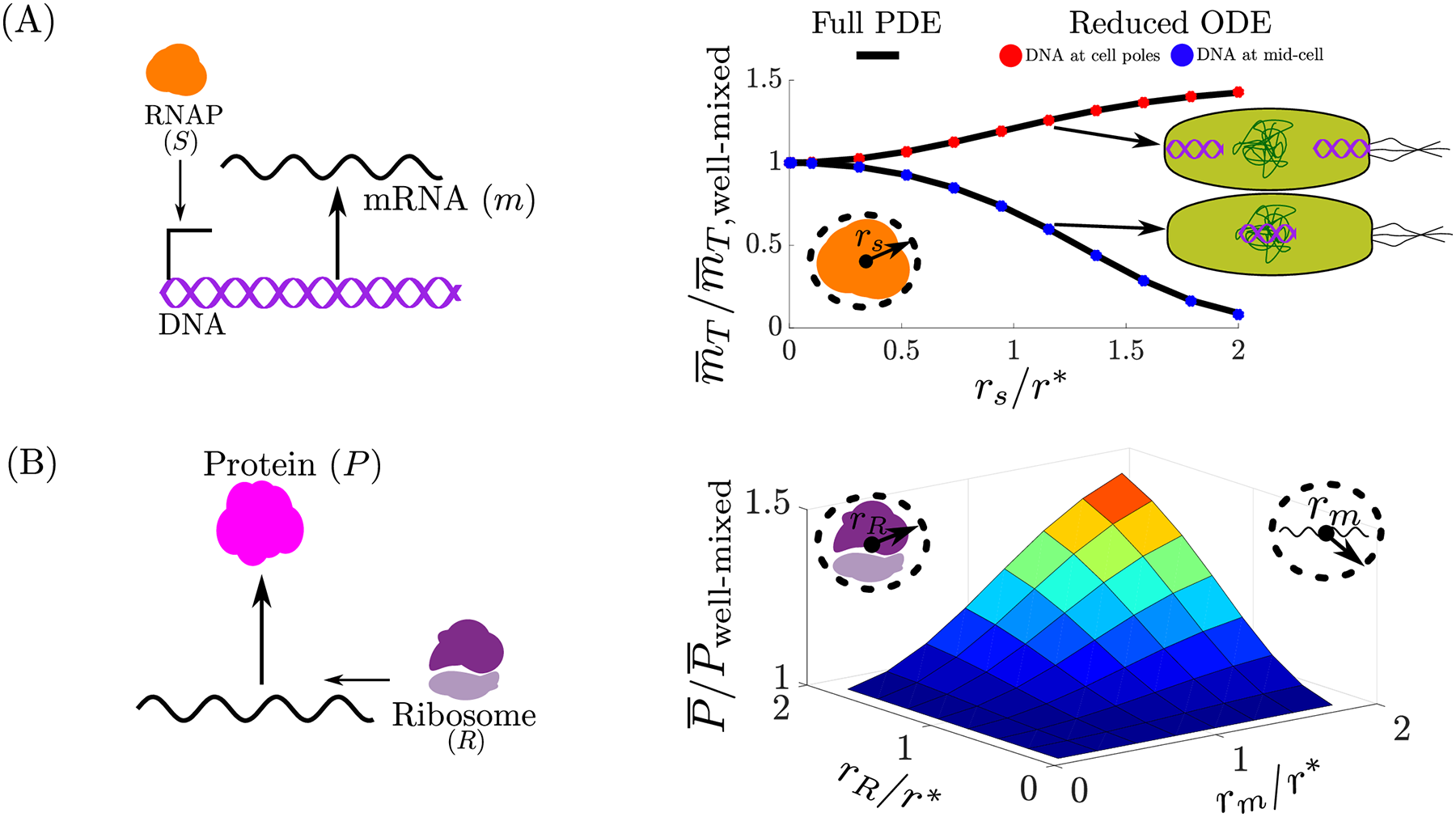
rates and reduced ODE model can capture these affects.
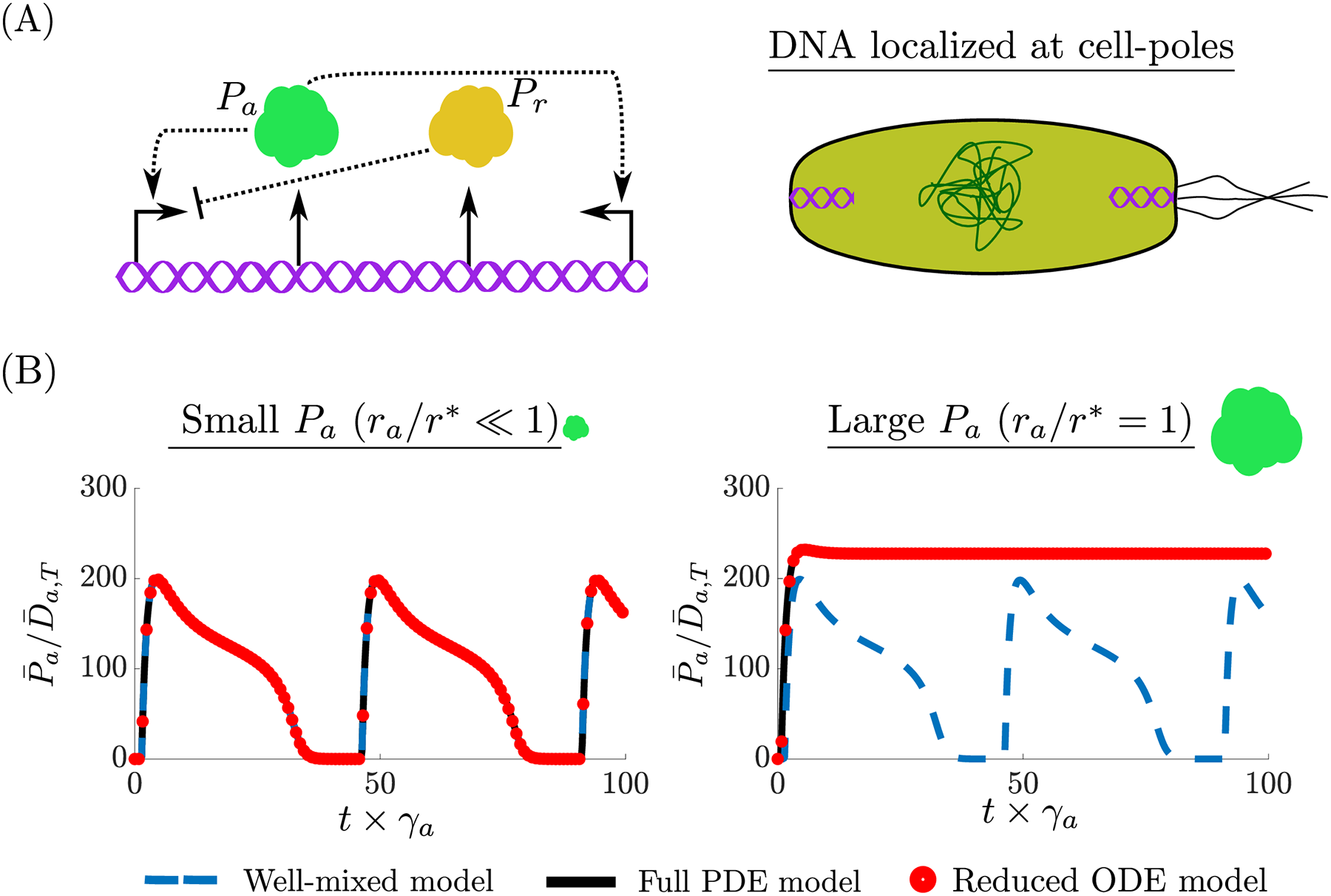
crucial in genetic circuit design
1. C. Barajas and D. Del Vecchio, “Effects of spatial heterogeneity on bacterial genetic circuits,” PLoS Comput. Biol., vol. 16, no. 9, (2020, doi).
2. C. Barajas and D. Del Vecchio, “Spatial Heterogeneity in Bacterial Cells,” IFAC Wold Congress, (2020, doi).
3. C. Barajas and D. Del Vecchio, “Genetic circuit-host ribosome transactions: Diffusion-reaction model,” in Proceedings of the American Control Conference, (2019, doi).
Resource controller to mitigate burden effects
Gene overexpression causes a burden on ribosomes, which decreases the expression of other genes and growth rate. I engineered a genetic ribosome actuator that increases gene expression rate and growth rate by over 150% and 80%, respectively. The actuator expresses the hydrolysis domain of the SpoT enzyme (SpoTH) in a bacterial strain with high basal levels of ppGpp, thus increasing ribosome availability. I use this actuator in a feedforward control arrangement, wherein SpoTH is co- expressed with a gene of interest (GOI). Without SpoTH co-expression, activation of the GOI decreases the expression of a constitutive gene and growth rate by over 80% and 40% , respectively. By contrast, with SpoTH co-expression, the ribosome binding site (RBS) of SpoTH can be tuned to achieve inappreciable change of either the expression of the constitutive gene or growth rate when the GOI is activated to the same level. We finally employ the actuator to keep growth rate constant while expressing dCas9 to a level that would otherwise decrease growth rate by more than 40% due to toxicity. This actuator will be relevant in applications where high cellular protein concentrations are desired without growth defects or global reduction of gene expression
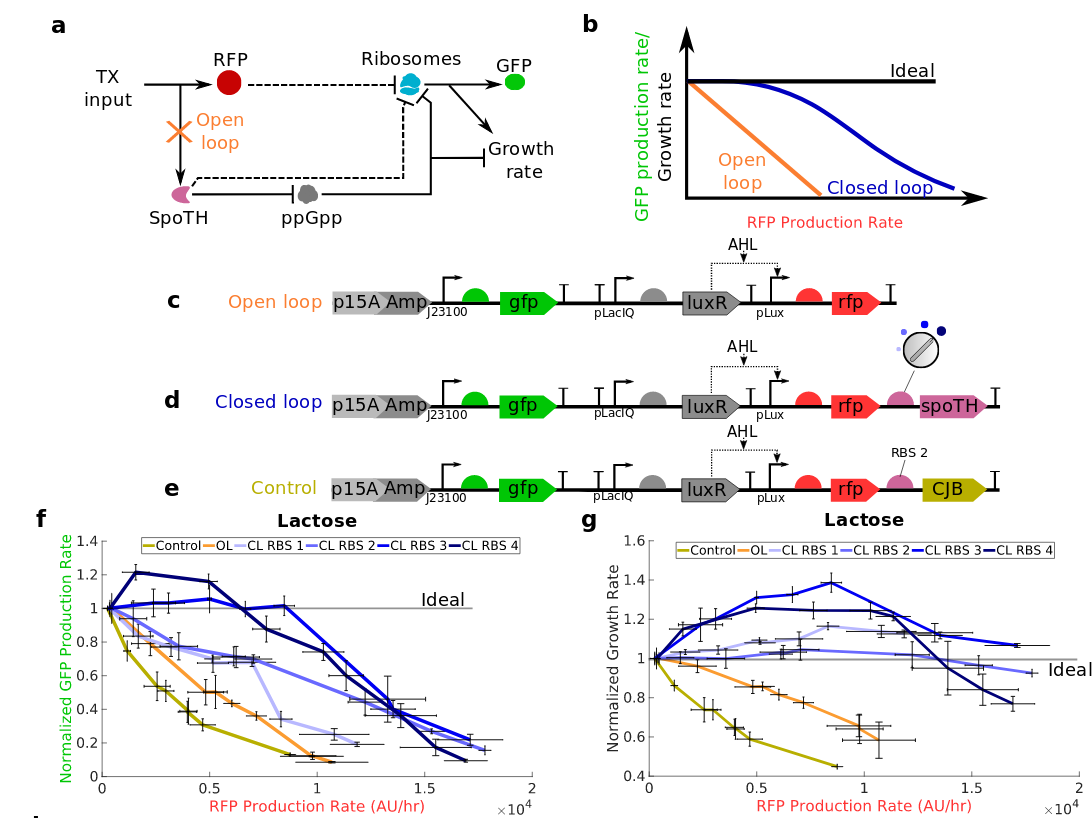
burden on gene expression and growth
rate due to overexpressing a heterologous
protein.
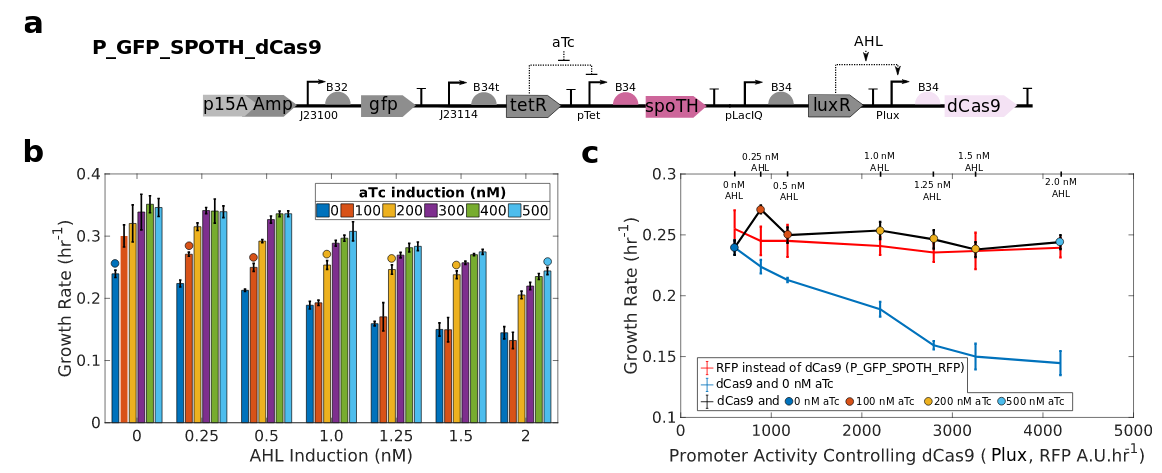
without growth defects.
Relevant publications:
1. C. Barajas , J. Gibson, L. Sandoval, and D. Del Vecchio, “Ribosome actuator via the SpoT/ppGpp pathway to mitigate gene overexpression burden,” bioRxiv, p. 2021.02.11.430724, (2021, doi).
Adaptive control and learning in biology
Recent technology leverages optogenetics to actuate gene expression and fluorescent proteins as sensors for gene activity. Thus allowing for precise control of gene expression. However, system parameters and model structure are typically unknown in gene expression networks, making them difficult to control. As part of my 2.153 project at MIT, I developed an adaptive control strategy that allows for transient tracking of a desired trajectory for protein expression in the presence of unknown parameters and nonlinearities. Not only, does the controller guarantee tracking, but given sufficient richness in the desired trajectory, It can learn model parameters and nonlinearities.
Left is tracking, middle is parameter that is learned, and right is nonliearity that is learned (direct link to video):